SQL for Data Scientists in 100 Queries
☆ what this is
- notes and working examples that instructors can use to perform a lesson
- do not expect novices with no prior SQL experience to be able to learn from them
- musical analogy
- this is the chord changes and melody
- we expect instructors to create an arrangement and/or improvise while delivering
- see Teaching Tech Together for background
☆ scope
- intended audience
- Rachel has a master’s degree in cell biology and now works in a research hospital doing cell assays.
- She learned a bit of R in an undergrad biostatistics course and has been through the Carpentries lesson on the Unix shell.
- Rachel is thinking about becoming a data scientist and would like to understand how data is stored and managed.
- Her work schedule is unpredictable and highly variable, so she needs to be able to learn a bit at a time.
- prerequisites
- basic Unix command line:
cd,ls,*wildcard - basic tabular data analysis: filtering rows, aggregating within groups
- basic Unix command line:
- learning outcomes
- Explain the difference between a database and a database manager.
- Write SQL to select, filter, sort, group, and aggregate data.
- Define tables and insert, update, and delete records.
- Describe different types of join and write queries that use them to combine data.
- Use windowing functions to operate on adjacent rows.
- Explain what transactions are and write queries that roll back when constraints are violated.
- Explain what triggers are and write SQL to create them.
- Manipulate JSON data using SQL.
- Interact with a database using Python directly, from a Jupyter notebook, and via an ORM.
☆ setup
- Download the latest release
- Unzip the file in a temporary directory to create:
./db/*.db: the SQLite databases used in the examples./src/*.*: SQL queries, Python scripts, and other source code./out/*.*: expected output for examples
☆ background concepts
- A database is a collection of data that can be searched and retrieved
- A database management system (DBMS) is a program that manages a particular kind of database
- Each DBMS stores data in its own way
- SQLite stores each database in a single file
- PostgreSQL spreads information across many files for higher performance
- DBMS can be a library embedded in other programs (SQLite) or a server (PostgreSQL)
- A relational database management system (RDBMS) stores data in tables and uses SQL for queries
- Unfortunately, every RDBMS has its own dialect of SQL
- There are also NoSQL databases like MongoDB that don’t use tables
☆ connect to database
sqlite3 data/penguins.db
- Not actually a query
- But we have to do it before we can do anything else
1: select constant
select 1;
1
selectis a keyword- Normally used to select data from table…
- …but if all we want is a constant value, we don’t need to specify one
- Semi-colon terminator is required
2: select all values from table
select * from little_penguins;
Adelie|Dream|37.2|18.1|178|3900|MALE
Adelie|Dream|37.6|19.3|181|3300|FEMALE
Gentoo|Biscoe|50|15.3|220|5550|MALE
Adelie|Torgersen|37.3|20.5|199|3775|MALE
Adelie|Biscoe|39.6|17.7|186|3500|FEMALE
Gentoo|Biscoe|47.7|15|216|4750|FEMALE
Adelie|Dream|36.5|18|182|3150|FEMALE
Gentoo|Biscoe|42|13.5|210|4150|FEMALE
Adelie|Torgersen|42.1|19.1|195|4000|MALE
Gentoo|Biscoe|54.3|15.7|231|5650|MALE
- An actual query
- Use
*to mean “all columns” - Use
from tablenameto specify table - Output format is not particularly readable
☆ administrative commands
.headers on
.mode markdown
select * from little_penguins;
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex |
|---------|-----------|----------------|---------------|-------------------|-------------|--------|
| Adelie | Dream | 37.2 | 18.1 | 178 | 3900 | MALE |
| Adelie | Dream | 37.6 | 19.3 | 181 | 3300 | FEMALE |
| Gentoo | Biscoe | 50 | 15.3 | 220 | 5550 | MALE |
| Adelie | Torgersen | 37.3 | 20.5 | 199 | 3775 | MALE |
| Adelie | Biscoe | 39.6 | 17.7 | 186 | 3500 | FEMALE |
| Gentoo | Biscoe | 47.7 | 15 | 216 | 4750 | FEMALE |
| Adelie | Dream | 36.5 | 18 | 182 | 3150 | FEMALE |
| Gentoo | Biscoe | 42 | 13.5 | 210 | 4150 | FEMALE |
| Adelie | Torgersen | 42.1 | 19.1 | 195 | 4000 | MALE |
| Gentoo | Biscoe | 54.3 | 15.7 | 231 | 5650 | MALE |
- SQLite administrative commands start with
.and aren’t part of the SQL standard- PostgreSQL’s special commands start with
\
- PostgreSQL’s special commands start with
- Use
.helpfor a complete list
3: specify columns
select species, island, sex
from little_penguins;
| species | island | sex |
|---------|-----------|--------|
| Adelie | Dream | MALE |
| Adelie | Dream | FEMALE |
| Gentoo | Biscoe | MALE |
| Adelie | Torgersen | MALE |
| Adelie | Biscoe | FEMALE |
| Gentoo | Biscoe | FEMALE |
| Adelie | Dream | FEMALE |
| Gentoo | Biscoe | FEMALE |
| Adelie | Torgersen | MALE |
| Gentoo | Biscoe | MALE |
- Specify column names separated by commas
- In any order
- Duplicates allowed
- Line breaks
allowedencouraged for readability
4: sort
select species, sex, island
from little_penguins
order by island asc, sex desc;
| species | sex | island |
|---------|--------|-----------|
| Gentoo | MALE | Biscoe |
| Gentoo | MALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
| Adelie | MALE | Dream |
| Adelie | FEMALE | Dream |
| Adelie | FEMALE | Dream |
| Adelie | MALE | Torgersen |
| Adelie | MALE | Torgersen |
order bymust followfrom(which must followselect)ascis ascending,descis descending- Default is ascending, but please specify
5: limit output
- Full dataset has 344 rows
select species, sex, island
from penguins
order by species, sex, island
limit 10;
| species | sex | island |
|---------|--------|-----------|
| Adelie | | Dream |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
- Comments start with
--and run to the end of the line limit Nspecifies maximum number of rows returned by query
6: page output
select species, sex, island
from penguins
order by species, sex, island
limit 10 offset 3;
| species | sex | island |
|---------|--------|-----------|
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
| Adelie | FEMALE | Biscoe |
offset Nmust followlimit- Specifies number of rows to skip from the start of the selection
- So this query skips the first 3 and shows the next 10
7: remove duplicates
select distinct species, sex, island
from penguins;
| species | sex | island |
|-----------|--------|-----------|
| Adelie | MALE | Torgersen |
| Adelie | FEMALE | Torgersen |
| Adelie | | Torgersen |
| Adelie | FEMALE | Biscoe |
| Adelie | MALE | Biscoe |
| Adelie | FEMALE | Dream |
| Adelie | MALE | Dream |
| Adelie | | Dream |
| Chinstrap | FEMALE | Dream |
| Chinstrap | MALE | Dream |
| Gentoo | FEMALE | Biscoe |
| Gentoo | MALE | Biscoe |
| Gentoo | | Biscoe |
distinctkeyword must appear right afterselect- SQL was supposed to read like English
- Shows distinct combinations
- Blanks in
sexcolumn show missing data- We’ll talk about this in a bit
8: filter results
select distinct species, sex, island
from penguins
where island = 'Biscoe';
| species | sex | island |
|---------|--------|--------|
| Adelie | FEMALE | Biscoe |
| Adelie | MALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
| Gentoo | MALE | Biscoe |
| Gentoo | | Biscoe |
where conditionfilters the rows produced by selection- Condition is evaluated independently for each row
- Only rows that pass the test appear in results
- Use single quotes for
'text data'and double quotes for"weird column names"- SQLite will accept double-quoted text data
9: filter with more complex conditions
select distinct species, sex, island
from penguins
where island = 'Biscoe' and sex != 'MALE';
| species | sex | island |
|---------|--------|--------|
| Adelie | FEMALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
and: both sub-conditions must be trueor: either or both part must be true- Notice that the row for Gentoo penguins on Biscoe island with unknown (empty) sex didn’t pass the test
- We’ll talk about this in a bit
10: do calculations
select
flipper_length_mm / 10.0,
body_mass_g / 1000.0
from penguins
limit 3;
| flipper_length_mm / 10.0 | body_mass_g / 1000.0 |
|--------------------------|----------------------|
| 18.1 | 3.75 |
| 18.6 | 3.8 |
| 19.5 | 3.25 |
- Can do the usual kinds of arithmetic on individual values
- Calculation done for each row independently
- Column name shows the calculation done
11: rename columns
select
flipper_length_mm / 10.0 as flipper_cm,
body_mass_g / 1000.0 as weight_kg,
island as where_found
from penguins
limit 3;
| flipper_cm | weight_kg | where_found |
|------------|-----------|-------------|
| 18.1 | 3.75 | Torgersen |
| 18.6 | 3.8 | Torgersen |
| 19.5 | 3.25 | Torgersen |
- Use
expression as nameto rename - Give result of calculation a meaningful name
- Can also rename columns without modifying
☆ check your understanding
12: calculate with missing values
select
flipper_length_mm / 10.0 as flipper_cm,
body_mass_g / 1000.0 as weight_kg,
island as where_found
from penguins
limit 5;
| flipper_cm | weight_kg | where_found |
|------------|-----------|-------------|
| 18.1 | 3.75 | Torgersen |
| 18.6 | 3.8 | Torgersen |
| 19.5 | 3.25 | Torgersen |
| | | Torgersen |
| 19.3 | 3.45 | Torgersen |
- SQL uses a special value
nullto representing missing data- Not 0 or empty string, but “I don’t know”
- Flipper length and body weight not known for one of the first five penguins
- “I don’t know” divided by 10 or 1000 is “I don’t know”
13: null equality
- Repeated from above so it doesn’t count against our query limit
select distinct species, sex, island
from penguins
where island = 'Biscoe';
| species | sex | island |
|---------|--------|--------|
| Adelie | FEMALE | Biscoe |
| Adelie | MALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
| Gentoo | MALE | Biscoe |
| Gentoo | | Biscoe |
- If we ask for female penguins the row with the missing sex drops out
select distinct species, sex, island
from penguins
where island = 'Biscoe' and sex = 'FEMALE';
| species | sex | island |
|---------|--------|--------|
| Adelie | FEMALE | Biscoe |
| Gentoo | FEMALE | Biscoe |
14: null inequality
- But if we ask for penguins that aren’t female it drops out as well
select distinct species, sex, island
from penguins
where island = 'Biscoe' and sex != 'FEMALE';
| species | sex | island |
|---------|------|--------|
| Adelie | MALE | Biscoe |
| Gentoo | MALE | Biscoe |
15: ternary logic
select null = null;
| null = null |
|-------------|
| |
- If we don’t know the left and right values, we don’t know if they’re equal or not
- So the result is
null - Get the same answer for
null != null - Ternary logic
| equality | |||
|---|---|---|---|
| X | Y | null | |
| X | true | false | null |
| Y | false | true | null |
| null | null | null | null |
16: handle null safely
select species, sex, island
from penguins
where sex is null;
| species | sex | island |
|---------|-----|-----------|
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Torgersen |
| Adelie | | Dream |
| Gentoo | | Biscoe |
| Gentoo | | Biscoe |
| Gentoo | | Biscoe |
| Gentoo | | Biscoe |
| Gentoo | | Biscoe |
- Use
is nullandis not nullto handle null safely - Other parts of SQL handle nulls specially
☆ check your understanding
17: aggregate
select sum(body_mass_g) as total_mass
from penguins;
| total_mass |
|------------|
| 1437000 |
- Aggregation combines many values to produce one
sumis an aggregation function- Combines corresponding values from multiple rows
18: common aggregation functions
select
max(bill_length_mm) as longest_bill,
min(flipper_length_mm) as shortest_flipper,
avg(bill_length_mm) / avg(bill_depth_mm) as weird_ratio
from penguins;
| longest_bill | shortest_flipper | weird_ratio |
|--------------|------------------|------------------|
| 59.6 | 172 | 2.56087082530644 |
- This actually shouldn’t work: can’t calculate maximum or average if any values are null
- SQL does the useful thing instead of the right one
19: counting
select
count(*) as count_star,
count(sex) as count_specific,
count(distinct sex) as count_distinct
from penguins;
| count_star | count_specific | count_distinct |
|------------|----------------|----------------|
| 344 | 333 | 2 |
count(*)counts rowscount(column)counts non-null entries in columncount(distinct column)counts distinct non-null entries
20: group
select
avg(body_mass_g) as average_mass_g
from penguins
group by sex;
| average_mass_g |
|------------------|
| 4005.55555555556 |
| 3862.27272727273 |
| 4545.68452380952 |
- Put rows in groups based on distinct combinations of values in columns specified with
group by - Then perform aggregation separately for each group
- But which is which?
21: behavior of unaggregated columns
select
sex,
avg(body_mass_g) as average_mass_g
from penguins
group by sex;
| sex | average_mass_g |
|--------|------------------|
| | 4005.55555555556 |
| FEMALE | 3862.27272727273 |
| MALE | 4545.68452380952 |
- All rows in each group have the same value for
sex, so no need to aggregate
22: arbitrary choice in aggregation
src/arbitrary_in_aggregation.sql
select
sex,
body_mass_g
from penguins
group by sex;
out/arbitrary_in_aggregation.out
| sex | body_mass_g |
|--------|-------------|
| | |
| FEMALE | 3800 |
| MALE | 3750 |
- If we don’t specify how to aggregate a column, SQL can choose any arbitrary value from the group
- All penguins in each group have the same sex because we grouped by that, so we get the right answer
- The body mass values are in the data but unpredictable
- A common mistake
23: filter aggregated values
select
sex,
avg(body_mass_g) as average_mass_g
from penguins
group by sex
having average_mass_g > 4000.0;
| sex | average_mass_g |
|------|------------------|
| | 4005.55555555556 |
| MALE | 4545.68452380952 |
- Using
having conditioninstead ofwhere conditionfor aggregates
24: readable output
select
sex,
round(avg(body_mass_g), 1) as average_mass_g
from penguins
group by sex
having average_mass_g > 4000.0;
| sex | average_mass_g |
|------|----------------|
| | 4005.6 |
| MALE | 4545.7 |
- Use
round(value, decimals)to round off a number
25: filter aggregate inputs
src/filter_aggregate_inputs.sql
select
sex,
round(
avg(body_mass_g) filter (where body_mass_g < 4000.0),
1
) as average_mass_g
from penguins
group by sex;
out/filter_aggregate_inputs.out
| sex | average_mass_g |
|--------|----------------|
| | 3362.5 |
| FEMALE | 3417.3 |
| MALE | 3752.5 |
filter (where condition)applies to inputs
☆ check your understanding
☆ create in-memory database
sqlite3 :memory:
- “Connect” to an in-memory database
26: create tables
create table job (
name text not null,
billable real not null
);
create table work (
person text not null,
job text not null
);
create table namefollowed by parenthesized list of columns- Each column is a name, a data type, and optional extra information
- E.g.,
not nullprevents nulls from being added
- E.g.,
.schemais not standard SQL- SQLite has added a few things
create if not exists- upper-case keywords (SQL is case insensitive)
27: insert data
insert into job values
('calibrate', 1.5),
('clean', 0.5);
insert into work values
('mik', 'calibrate'),
('mik', 'clean'),
('mik', 'complain'),
('po', 'clean'),
('po', 'complain'),
('tay', 'complain');
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
| clean | 0.5 |
| person | job |
|--------|-----------|
| mik | calibrate |
| mik | clean |
| mik | complain |
| po | clean |
| po | complain |
| tay | complain |
28: update rows
update work
set person = "tae"
where person = "tay";
| person | job |
|--------|-----------|
| mik | calibrate |
| mik | clean |
| mik | complain |
| po | clean |
| po | complain |
| tae | complain |
- (Almost) always specify row(w) to update using
where- Would otherwise update all rows
- Useful to give each row a primary key that uniquely identifies it for this purpose
- Will see other uses below
29: delete rows
delete from work
where person = "tae";
select * from work;
| person | job |
|--------|-----------|
| mik | calibrate |
| mik | clean |
| mik | complain |
| po | clean |
| po | complain |
- Again, (almost) always specify row(s) to delete using
where
30: backing up
create table backup (
person text not null,
job text not null
);
insert into backup
select person, job
from work
where person = 'tae';
delete from work
where person = 'tae';
select * from backup;
| person | job |
|--------|----------|
| tae | complain |
☆ check your understanding
31: join tables
select *
from work cross join job;
| person | job | name | billable |
|--------|-----------|-----------|----------|
| mik | calibrate | calibrate | 1.5 |
| mik | calibrate | clean | 0.5 |
| mik | clean | calibrate | 1.5 |
| mik | clean | clean | 0.5 |
| mik | complain | calibrate | 1.5 |
| mik | complain | clean | 0.5 |
| po | clean | calibrate | 1.5 |
| po | clean | clean | 0.5 |
| po | complain | calibrate | 1.5 |
| po | complain | clean | 0.5 |
| tay | complain | calibrate | 1.5 |
| tay | complain | clean | 0.5 |
- A join combines information from two tables
- full outer join (also called cross join) constructs their cross product
- All combinations of rows from each
- Result isn’t particularly useful:
jobandnamedon’t match
32: inner join
select *
from work inner join job
on work.job = job.name;
| person | job | name | billable |
|--------|-----------|-----------|----------|
| mik | calibrate | calibrate | 1.5 |
| mik | clean | clean | 0.5 |
| po | clean | clean | 0.5 |
- Use
table.columnnotation to specify columns- A column can have the same name as a table
- Use
on conditionto specify join condition - Since
complaindoesn’t appear injob.name, none of those rows are kept
33: aggregate joined data
select
work.person,
sum(job.billable) as pay
from work inner join job
on work.job = job.name
group by work.person;
| person | pay |
|--------|-----|
| mik | 2.0 |
| po | 0.5 |
- Combines ideas we’ve seen before
- But Tay is missing from the table
34: left join
select *
from work left join job
on work.job = job.name;
| person | job | name | billable |
|--------|-----------|-----------|----------|
| mik | calibrate | calibrate | 1.5 |
| mik | clean | clean | 0.5 |
| mik | complain | | |
| po | clean | clean | 0.5 |
| po | complain | | |
| tay | complain | | |
- A left outer join keeps all rows from the left table
- Fills missing values from right table with null
35: aggregate left joins
select
work.person,
sum(job.billable) as pay
from work left join job
on work.job = job.name
group by work.person;
| person | pay |
|--------|-----|
| mik | 2.0 |
| po | 0.5 |
| tay | |
- That’s better, but we’d like to see 0 rather than a blank
☆ check your understanding
36: coalesce values
select
work.person,
coalesce(sum(job.billable), 0.0) as pay
from work left join job
on work.job = job.name
group by work.person;
| person | pay |
|--------|-----|
| mik | 2.0 |
| po | 0.5 |
| tay | 0.0 |
coalesce(val1, val2, …)returns first non-null value
37: negate incorrectly
- Who doesn’t calibrate?
select distinct person
from work
where job != 'calibrate';
| person |
|--------|
| mik |
| po |
| tay |
- But Mik does calibrate
- Problem is that there’s an entry for Mik cleaning
- And since
'clean' != 'calibrate', that row is included in the results - We need a different approach
38: set membership
select *
from work
where person not in ('mik', 'tay');
| person | job |
|--------|----------|
| po | clean |
| po | complain |
in valuesandnot in valuesdo exactly what you expect
39: subqueries
select distinct person
from work
where person not in (
select distinct person
from work
where job = 'calibrate'
);
| person |
|--------|
| po |
| tay |
- Use a subquery to select the people who do calibrate
- Then select all the people who aren’t in that set
- Initially feels odd, but subqueries are useful in other ways
☆ M to N relationships
- Relationships between entities are usually characterized as:
- 1-to-1: fields in the same record
- 1-to-many: the many have a foreign key referring to the one’s primary key
- many-to-many: don’t know how many keys to add to records (“maximum” never is)
- Nearly-universal solution is a join table
- Each record is a pair of foreign keys
- I.e., each record is the fact that records A and B are related
40: autoincrement and primary key
create table person (
ident integer primary key autoincrement,
name text not null
);
insert into person values
(null, 'mik'),
(null, 'po'),
(null, 'tay');
select * from person;
insert into person values (1, "prevented");
| ident | name |
|-------|------|
| 1 | mik |
| 2 | po |
| 3 | tay |
Runtime error near line 12: UNIQUE constraint failed: person.ident (19)
- Database autoincrements
identeach time a new record is added - Use that field as the primary key
- So that if Mik changes their name again, we only have to change one fact in the database
- Downside: manual queries are harder to read (who is person 17?)
☆ internal tables
select * from sqlite_sequence;
| name | seq |
|--------|-----|
| person | 3 |
- Sequence numbers are not reset when rows are deleted
41: alter tables
alter table job
add ident integer not null default -1;
update job
set ident = 1
where name = 'calibrate';
update job
set ident = 2
where name = 'clean';
select * from job;
| name | billable | ident |
|-----------|----------|-------|
| calibrate | 1.5 | 1 |
| clean | 0.5 | 2 |
- Add a column after the fact
- Since it can’t be null, we have to provide a default value
- Really want to make it the primary key, but SQLite doesn’t allow that (easily) after the fact
- Then use
updateto modify existing records- Can modify any number of records at once
- So be careful about
whereclause
- Data migration
42: create new tables from old
create table new_work (
person_id integer not null,
job_id integer not null,
foreign key(person_id) references person(ident),
foreign key(job_id) references job(ident)
);
insert into new_work
select
person.ident as person_id,
job.ident as job_id
from
(person join work on person.name = work.person)
join job on job.name = work.job;
select * from new_work;
| person_id | job_id |
|-----------|--------|
| 1 | 1 |
| 1 | 2 |
| 2 | 2 |
new_workis our join table- Each column refers to a record in some other table
43: remove tables
drop table work;
alter table new_work rename to work;
CREATE TABLE job (
ident integer primary key autoincrement,
name text not null,
billable real not null
);
CREATE TABLE sqlite_sequence(name,seq);
CREATE TABLE person (
ident integer primary key autoincrement,
name text not null
);
CREATE TABLE IF NOT EXISTS "work" (
person_id integer not null,
job_id integer not null,
foreign key(person_id) references person(ident),
foreign key(job_id) references job(ident)
);
- Remove the old table and rename the new one to take its place
- Note
if exists
- Note
- Be careful…
44: compare individual values to aggregates
- Go back to penguins
src/compare_individual_aggregate.sql
select body_mass_g
from penguins
where body_mass_g > (
select avg(body_mass_g)
from penguins
)
limit 5;
out/compare_individual_aggregate.out
| body_mass_g |
|-------------|
| 4675 |
| 4250 |
| 4400 |
| 4500 |
| 4650 |
- Get average body mass in subquery
- Compare each row against that
- Requires two scans of the data, but there’s no way to avoid that
- Null values aren’t included in the average or in the final results
45: compare individual values to aggregates within groups
select
penguins.species,
penguins.body_mass_g,
round(averaged.avg_mass_g, 1) as avg_mass_g
from penguins join (
select species, avg(body_mass_g) as avg_mass_g
from penguins
group by species
) as averaged
on penguins.species = averaged.species
where penguins.body_mass_g > averaged.avg_mass_g
limit 5;
| species | body_mass_g | avg_mass_g |
|---------|-------------|------------|
| Adelie | 3750 | 3700.7 |
| Adelie | 3800 | 3700.7 |
| Adelie | 4675 | 3700.7 |
| Adelie | 4250 | 3700.7 |
| Adelie | 3800 | 3700.7 |
- Subquery runs first to create temporary table
averagedwith average mass per species - Join that with
penguins - Filter to find penguins heavier than average within their species
46: common table expressions
src/common_table_expressions.sql
with grouped as (
select species, avg(body_mass_g) as avg_mass_g
from penguins
group by species
)
select
penguins.species,
penguins.body_mass_g,
round(grouped.avg_mass_g, 1) as avg_mass_g
from penguins join grouped
where penguins.body_mass_g > grouped.avg_mass_g
limit 5;
out/common_table_expressions.out
| species | body_mass_g | avg_mass_g |
|---------|-------------|------------|
| Adelie | 3750 | 3700.7 |
| Adelie | 3800 | 3700.7 |
| Adelie | 4675 | 3700.7 |
| Adelie | 4250 | 3700.7 |
| Adelie | 3800 | 3700.7 |
- Use common table expression (CTE) to make queries clearer
- Nested subqueries quickly become difficult to understand
- Database decides how to optimize
☆ explain query plan
explain query plan
select
species,
avg(body_mass_g)
from penguins
group by species;
QUERY PLAN
|--SCAN penguins
`--USE TEMP B-TREE FOR GROUP BY
- SQLite plans to scan every row of the table
- It will build a temporary B-tree data structure to group rows
47: enumerate rows
- Every table has a special column called
rowid
select rowid, species, island
from penguins
limit 5;
| rowid | species | island |
|-------|---------|-----------|
| 1 | Adelie | Torgersen |
| 2 | Adelie | Torgersen |
| 3 | Adelie | Torgersen |
| 4 | Adelie | Torgersen |
| 5 | Adelie | Torgersen |
rowidis persistent within a session- I.e., if we delete the first 5 rows we now have row IDs 6…N
- Do not rely on row ID
- In particular, do not use it as a key
48: if-else function
with sized_penguins as (
select
species,
iif(
body_mass_g < 3500,
'small',
'large'
) as size
from penguins
)
select species, size, count(*) as num
from sized_penguins
group by species, size
order by species, num;
| species | size | num |
|-----------|-------|-----|
| Adelie | small | 54 |
| Adelie | large | 98 |
| Chinstrap | small | 17 |
| Chinstrap | large | 51 |
| Gentoo | large | 124 |
iif(condition, true_result, false_result)- Note:
iifwith two i’s
- Note:
49: select a case
- What if we want small, medium, and large?
- Can nest
iif, but quickly becomes unreadable
with sized_penguins as (
select
species,
case
when body_mass_g < 3500 then 'small'
when body_mass_g < 5000 then 'medium'
else 'large'
end as size
from penguins
)
select species, size, count(*) as num
from sized_penguins
group by species, size
order by species, num;
| species | size | num |
|-----------|--------|-----|
| Adelie | large | 1 |
| Adelie | small | 54 |
| Adelie | medium | 97 |
| Chinstrap | small | 17 |
| Chinstrap | medium | 51 |
| Gentoo | medium | 56 |
| Gentoo | large | 68 |
- Evaluate
whenoptions in order and take first - Result of
caseis null if no condition is true - Use
elseas fallback
50: check range
with sized_penguins as (
select
species,
case
when body_mass_g between 3500 and 5000 then 'normal'
else 'abnormal'
end as size
from penguins
)
select species, size, count(*) as num
from sized_penguins
group by species, size
order by species, num;
| species | size | num |
|-----------|----------|-----|
| Adelie | abnormal | 55 |
| Adelie | normal | 97 |
| Chinstrap | abnormal | 17 |
| Chinstrap | normal | 51 |
| Gentoo | abnormal | 62 |
| Gentoo | normal | 62 |
betweencan make queries easier to read- But be careful of the
andin the middle
☆ yet another database
- Entity-relationship diagram (ER diagram) shows relationships between tables
- Like everything to do with databases, there are lots of variations
select * from staff;
| ident | personal | family | dept | age |
|-------|----------|-----------|------|-----|
| 1 | Kartik | Gupta | | 46 |
| 2 | Divit | Dhaliwal | hist | 34 |
| 3 | Indrans | Sridhar | mb | 47 |
| 4 | Pranay | Khanna | mb | 51 |
| 5 | Riaan | Dua | | 23 |
| 6 | Vedika | Rout | hist | 45 |
| 7 | Abram | Chokshi | gen | 23 |
| 8 | Romil | Kapoor | hist | 38 |
| 9 | Ishaan | Ramaswamy | mb | 35 |
| 10 | Nitya | Lal | gen | 52 |
51: pattern matching
select personal, family from staff
where personal like '%ya%' or family glob '*De*';
| personal | family |
|----------|--------|
| Nitya | Lal |
likeis the original SQL pattern matcher%matches zero or more characters at the start or end of a string- Case insensitive by default
globsupports Unix-style wildcards
| name | purpose |
|---|---|
substr |
Get substring given starting point and length |
trim |
Remove characters from beginning and end of string |
ltrim |
Remove characters from beginning of string |
rtrim |
Remove characters from end of string |
length |
Length of string |
replace |
Replace occurrences of substring with another string |
upper |
Return upper-case version of string |
lower |
Return lower-case version of string |
instr |
Find location of first occurrence of substring (returns 0 if not found) |
52: select first and last rows
select * from (
select * from (select * from experiment order by started asc limit 5)
union all
select * from (select * from experiment order by started desc limit 5)
)
order by started asc
;
| ident | kind | started | ended |
|-------|-------------|------------|------------|
| 17 | trial | 2023-01-29 | 2023-01-30 |
| 35 | calibration | 2023-01-30 | 2023-01-30 |
| 36 | trial | 2023-02-02 | 2023-02-03 |
| 25 | trial | 2023-02-12 | 2023-02-14 |
| 2 | calibration | 2023-02-14 | 2023-02-14 |
| 40 | calibration | 2024-01-21 | 2024-01-21 |
| 12 | trial | 2024-01-26 | 2024-01-28 |
| 44 | trial | 2024-01-27 | 2024-01-29 |
| 34 | trial | 2024-02-01 | 2024-02-02 |
| 14 | calibration | 2024-02-03 | 2024-02-03 |
union allcombines records- Keeps duplicates:
unionon its own keeps unique records
- Keeps duplicates:
- Yes, it feels like the extra
select * fromshould be unnecessary
53: intersection
select personal, family, dept, age
from staff
where dept = 'mb'
intersect
select personal, family, dept, age from staff
where age < 50
;
| personal | family | dept | age |
|----------|-----------|------|-----|
| Indrans | Sridhar | mb | 47 |
| Ishaan | Ramaswamy | mb | 35 |
- Tables being intersected must have same structure
- Intersection usually used when pulling values from different tables
- In this case, would be clearer to use
where
- In this case, would be clearer to use
54: exclusion
select personal, family, dept, age
from staff
where dept = 'mb'
except
select personal, family, dept, age from staff
where age < 50
;
| personal | family | dept | age |
|----------|--------|------|-----|
| Pranay | Khanna | mb | 51 |
- Again, tables must have same structure
- And this would be clearer with
where
- And this would be clearer with
- SQL operates on sets, not tables, except where it doesn’t
55: random numbers and why not
with decorated as (
select random() as rand,
personal || ' ' || family as name
from staff
)
select rand, abs(rand) % 10 as selector, name
from decorated
where selector < 5;
| rand | selector | name |
|----------------------|----------|-----------------|
| 7176652035743196310 | 0 | Divit Dhaliwal |
| -2243654635505630380 | 2 | Indrans Sridhar |
| -6940074802089166303 | 5 | Pranay Khanna |
| 8882650891091088193 | 9 | Riaan Dua |
| -45079732302991538 | 5 | Vedika Rout |
| -8973877087806386134 | 2 | Abram Chokshi |
| 3360598450426870356 | 9 | Romil Kapoor |
- There is no way to seed SQLite’s random number generator
- Which means there is no way to reproduce one of its “random” sequences
56: creating index
explain query plan
select filename
from plate
where filename like '%07%';
create index plate_file on plate(filename);
explain query plan
select filename
from plate
where filename like '%07%';
QUERY PLAN
`--SCAN plate USING COVERING INDEX sqlite_autoindex_plate_1
QUERY PLAN
`--SCAN plate USING COVERING INDEX plate_file
- An index is an auxiliary data structure that enables faster access to records
- Spend storage space to buy speed
- Don’t have to mention it explicitly in queries
- Database manager will use it automatically
57: generate sequence
select value from generate_series(1, 5);
| value |
|-------|
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
- A (non-standard) table-valued function
58: generate sequence based on data
create table temp (
num integer not null
);
insert into temp values (1), (5);
select value from generate_series (
(select min(num) from temp),
(select max(num) from temp)
);
| value |
|-------|
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
- Must have the parentheses around the
minandmaxselections to keep SQLite happy
59: generate sequence of dates
select
date((select julianday(min(started)) from experiment) + value) as some_day
from (
select value from generate_series(
(select 0),
(select count(*) - 1 from experiment)
)
)
limit 5;
| some_day |
|------------|
| 2023-01-29 |
| 2023-01-30 |
| 2023-01-31 |
| 2023-02-01 |
| 2023-02-02 |
- SQLite represents dates as YYYY-MM-DD strings
or as Julian days or as Unix milliseconds or…
- Julian days is fractional number of days since November 24, 4714 BCE
juliandayanddateconvert back and forth
60: count experiments started per day without gaps
with
-- complete sequence of days with 0 as placeholder for number of experiments
all_days as (
select
date((select julianday(min(started)) from experiment) + value) as some_day,
0 as zeroes
from (
select value from generate_series(
(select 0),
(select count(*) - 1 from experiment)
)
)
),
-- sequence of actual days with actual number of experiments started
actual_days as (
select started, count(started) as num_exp
from experiment
group by started
)
-- combined by joining on day and taking actual number (if available) or zero
select
all_days.some_day as day,
coalesce(actual_days.num_exp, all_days.zeroes) as num_exp
from
all_days left join actual_days on all_days.some_day = actual_days.started
limit 5
;
| day | num_exp |
|------------|---------|
| 2023-01-29 | 1 |
| 2023-01-30 | 1 |
| 2023-01-31 | 0 |
| 2023-02-01 | 0 |
| 2023-02-02 | 1 |
61: self join
with person as (
select
ident,
personal || ' ' || family as name
from staff
)
select left.name, right.name
from person as left join person as right
limit 10;
| name | name |
|--------------|------------------|
| Kartik Gupta | Kartik Gupta |
| Kartik Gupta | Divit Dhaliwal |
| Kartik Gupta | Indrans Sridhar |
| Kartik Gupta | Pranay Khanna |
| Kartik Gupta | Riaan Dua |
| Kartik Gupta | Vedika Rout |
| Kartik Gupta | Abram Chokshi |
| Kartik Gupta | Romil Kapoor |
| Kartik Gupta | Ishaan Ramaswamy |
| Kartik Gupta | Nitya Lal |
- Join a table to itself
- Use
asto create aliases for copies of tables to distinguish them - Nothing special about the names
leftandright
- Use
- Get all pairs, including person with themself
62: generate unique pairs
with person as (
select
ident,
personal || ' ' || family as name
from staff
)
select left.name, right.name
from person as left join person as right
on left.ident < right.ident
where left.ident <= 4 and right.ident <= 4;
| name | name |
|-----------------|-----------------|
| Kartik Gupta | Divit Dhaliwal |
| Kartik Gupta | Indrans Sridhar |
| Kartik Gupta | Pranay Khanna |
| Divit Dhaliwal | Indrans Sridhar |
| Divit Dhaliwal | Pranay Khanna |
| Indrans Sridhar | Pranay Khanna |
left.ident < right.identensures distinct pairs without duplicates- Use
left.ident <= 4 and right.ident <= 4to limit output - Quick check: pairs
63: filter pairs
with
person as (
select
ident,
personal || ' ' || family as name
from staff
),
together as (
select
left.staff as left_staff,
right.staff as right_staff
from performed as left join performed as right
on left.experiment = right.experiment
where left_staff < right_staff
)
select
left.name as person_1,
right.name as person_2
from person as left join person as right join together
on left.ident = left_staff and right.ident = right_staff;
| person_1 | person_2 |
|-----------------|------------------|
| Kartik Gupta | Vedika Rout |
| Pranay Khanna | Vedika Rout |
| Indrans Sridhar | Romil Kapoor |
| Abram Chokshi | Ishaan Ramaswamy |
| Pranay Khanna | Vedika Rout |
| Kartik Gupta | Abram Chokshi |
| Abram Chokshi | Romil Kapoor |
| Kartik Gupta | Divit Dhaliwal |
| Divit Dhaliwal | Abram Chokshi |
| Pranay Khanna | Ishaan Ramaswamy |
| Indrans Sridhar | Romil Kapoor |
| Kartik Gupta | Ishaan Ramaswamy |
| Kartik Gupta | Nitya Lal |
| Kartik Gupta | Abram Chokshi |
| Pranay Khanna | Romil Kapoor |
64: existence and correlated subqueries
select name, building
from department
where exists (
select 1
from staff
where dept = department.ident
)
order by name;
| name | building |
|-------------------|------------------|
| Genetics | Chesson |
| Histology | Fashet Extension |
| Molecular Biology | Chesson |
- Nobody works in Endocrinology
select 1could equally beselect trueor any other value- A correlated subquery depends on a value from the outer query
- Equivalent to nested loop
65: nonexistence
select name, building
from department
where not exists (
select 1
from staff
where dept = department.ident
)
order by name;
| name | building |
|---------------|----------|
| Endocrinology | TGVH |
☆ avoiding correlated subqueries
src/avoid_correlated_subqueries.sql
select distinct
department.name as name,
department.building as building
from department join staff
on department.ident = staff.dept
order by name;
out/avoid_correlated_subqueries.out
| name | building |
|-------------------|------------------|
| Genetics | Chesson |
| Histology | Fashet Extension |
| Molecular Biology | Chesson |
- The join might or might not be faster than the correlated subquery
- Hard to find unstaffed departments without either
not existsorcountand a check for 0
66: lead and lag
with ym_num as (
select
strftime('%Y-%m', started) as ym,
count(*) as num
from experiment
group by ym
)
select
ym,
lag(num) over (order by ym) as prev_num,
num,
lead(num) over (order by ym) as next_num
from ym_num
order by ym;
| ym | prev_num | num | next_num |
|---------|----------|-----|----------|
| 2023-01 | | 2 | 5 |
| 2023-02 | 2 | 5 | 5 |
| 2023-03 | 5 | 5 | 1 |
| 2023-04 | 5 | 1 | 6 |
| 2023-05 | 1 | 6 | 5 |
| 2023-06 | 6 | 5 | 3 |
| 2023-07 | 5 | 3 | 2 |
| 2023-08 | 3 | 2 | 4 |
| 2023-09 | 2 | 4 | 6 |
| 2023-10 | 4 | 6 | 4 |
| 2023-12 | 6 | 4 | 5 |
| 2024-01 | 4 | 5 | 2 |
| 2024-02 | 5 | 2 | |
- Use
strftimeto extract year and month- Clumsy, but date/time handling is not SQLite’s strong point
- Use window functions
leadandlagto shift values- Unavailable values are null
67: window functions
with ym_num as (
select
strftime('%Y-%m', started) as ym,
count(*) as num
from experiment
group by ym
)
select
ym,
num,
sum(num) over (order by ym) as num_done,
cume_dist() over (order by ym) as progress
from ym_num
order by ym;
| ym | num | num_done | progress |
|---------|-----|----------|--------------------|
| 2023-01 | 2 | 2 | 0.0769230769230769 |
| 2023-02 | 5 | 7 | 0.153846153846154 |
| 2023-03 | 5 | 12 | 0.230769230769231 |
| 2023-04 | 1 | 13 | 0.307692307692308 |
| 2023-05 | 6 | 19 | 0.384615384615385 |
| 2023-06 | 5 | 24 | 0.461538461538462 |
| 2023-07 | 3 | 27 | 0.538461538461538 |
| 2023-08 | 2 | 29 | 0.615384615384615 |
| 2023-09 | 4 | 33 | 0.692307692307692 |
| 2023-10 | 6 | 39 | 0.769230769230769 |
| 2023-12 | 4 | 43 | 0.846153846153846 |
| 2024-01 | 5 | 48 | 0.923076923076923 |
| 2024-02 | 2 | 50 | 1.0 |
sum() overdoes a running totalcume_distis fraction of rows seen so far
☆ explain another query plain
src/explain_window_function.sql
explain query plan
with ym_num as (
select
strftime('%Y-%m', started) as ym,
count(*) as num
from experiment
group by ym
)
select
ym,
num,
sum(num) over (order by ym) as num_done,
cume_dist() over (order by ym) as progress
from ym_num
order by ym;
out/explain_window_function.out
QUERY PLAN
|--CO-ROUTINE (subquery-3)
| |--CO-ROUTINE (subquery-4)
| | |--CO-ROUTINE ym_num
| | | |--SCAN experiment
| | | `--USE TEMP B-TREE FOR GROUP BY
| | |--SCAN ym_num
| | `--USE TEMP B-TREE FOR ORDER BY
| `--SCAN (subquery-4)
`--SCAN (subquery-3)
- Becomes useful…eventually
68: partitioned windows
with y_m_num as (
select
strftime('%Y', started) as year,
strftime('%m', started) as month,
count(*) as num
from experiment
group by year, month
)
select
year,
month,
num,
sum(num) over (partition by year order by month) as num_done
from y_m_num
order by year, month;
| year | month | num | num_done |
|------|-------|-----|----------|
| 2023 | 01 | 2 | 2 |
| 2023 | 02 | 5 | 7 |
| 2023 | 03 | 5 | 12 |
| 2023 | 04 | 1 | 13 |
| 2023 | 05 | 6 | 19 |
| 2023 | 06 | 5 | 24 |
| 2023 | 07 | 3 | 27 |
| 2023 | 08 | 2 | 29 |
| 2023 | 09 | 4 | 33 |
| 2023 | 10 | 6 | 39 |
| 2023 | 12 | 4 | 43 |
| 2024 | 01 | 5 | 5 |
| 2024 | 02 | 2 | 7 |
partition bycreates groups- So this counts experiments started since the beginning of each year
69: blobs
create table images (
name text not null,
content blob
);
insert into images(name, content) values
("biohazard", readfile("img/biohazard.png")),
("crush", readfile("img/crush.png")),
("fire", readfile("img/fire.png")),
("radioactive", readfile("img/radioactive.png")),
("tripping", readfile("img/tripping.png"));
select name, length(content) from images;
| name | length(content) |
|-------------|-----------------|
| biohazard | 19629 |
| crush | 15967 |
| fire | 18699 |
| radioactive | 16661 |
| tripping | 17208 |
☆ yet another database
sqlite3 data/lab_log.db
.schema
CREATE TABLE sqlite_sequence(name,seq);
CREATE TABLE person(
ident integer primary key autoincrement,
details text not null
);
CREATE TABLE machine(
ident integer primary key autoincrement,
name text not null,
details text not null
);
CREATE TABLE usage(
ident integer primary key autoincrement,
log text not null
);
70: store JSON
select * from machine;
| ident | name | details |
|-------|----------------|---------------------------------------------------------|
| 1 | WY401 | {"acquired": "2023-05-01"} |
| 2 | Inphormex | {"acquired": "2021-07-15", "refurbished": "2023-10-22"} |
| 3 | AutoPlate 9000 | {"note": "needs software update"} |
- Store heterogeneous data as JSON-formatted text (with double-quoted strings)
- Database parses it each time it is queried
- Alternatively store as blob
- Can’t just view it
- But more efficient
71: select field from JSON
select
details->'$.acquired' as single_arrow,
details->>'$.acquired' as double_arrow
from machine;
| single_arrow | double_arrow |
|--------------|--------------|
| "2023-05-01" | 2023-05-01 |
| "2021-07-15" | 2021-07-15 |
| | |
- Single arrow
->returns JSON representation result - Double arrow
->>returns SQL text, integer, real, or null - Left side is column
- Right side is path expression
- Start with
$(meaning “root”) - Fields separated by
.
- Start with
72: JSON array access
select
ident,
json_array_length(log->'$') as length,
log->'$[0]' as first
from usage;
| ident | length | first |
|-------|--------|--------------------------------------------------------------|
| 1 | 4 | {"machine":"Inphormex","person":["Gabrielle","Dub\u00e9"]} |
| 2 | 5 | {"machine":"Inphormex","person":["Marianne","Richer"]} |
| 3 | 2 | {"machine":"sterilizer","person":["Josette","Villeneuve"]} |
| 4 | 1 | {"machine":"sterilizer","person":["Maude","Goulet"]} |
| 5 | 2 | {"machine":"AutoPlate 9000","person":["Brigitte","Michaud"]} |
| 6 | 1 | {"machine":"sterilizer","person":["Marianne","Richer"]} |
| 7 | 3 | {"machine":"WY401","person":["Maude","Goulet"]} |
| 8 | 1 | {"machine":"AutoPlate 9000"} |
- SQLite (and other database managers) has lots of JSON manipulation functions
json_array_lengthgives number of elements in selected array- subscripts start with 0
- Characters outside 7-bit ASCII represented as Unicode escapes
73: unpack JSON array
select
ident,
json_each.key as key,
json_each.value as value
from usage, json_each(usage.log)
limit 10;
| ident | key | value |
|-------|-----|--------------------------------------------------------------|
| 1 | 0 | {"machine":"Inphormex","person":["Gabrielle","Dub\u00e9"]} |
| 1 | 1 | {"machine":"Inphormex","person":["Gabrielle","Dub\u00e9"]} |
| 1 | 2 | {"machine":"WY401","person":["Gabrielle","Dub\u00e9"]} |
| 1 | 3 | {"machine":"Inphormex","person":["Gabrielle","Dub\u00e9"]} |
| 2 | 0 | {"machine":"Inphormex","person":["Marianne","Richer"]} |
| 2 | 1 | {"machine":"AutoPlate 9000","person":["Marianne","Richer"]} |
| 2 | 2 | {"machine":"sterilizer","person":["Marianne","Richer"]} |
| 2 | 3 | {"machine":"AutoPlate 9000","person":["Monique","Marcotte"]} |
| 2 | 4 | {"machine":"sterilizer","person":["Marianne","Richer"]} |
| 3 | 0 | {"machine":"sterilizer","person":["Josette","Villeneuve"]} |
json_eachis another table-valued function- Use
json_each.nameto get properties of unpacked array
74: last element of array
select
ident,
log->'$[#-1].machine' as final
from usage
limit 5;
| ident | final |
|-------|--------------|
| 1 | "Inphormex" |
| 2 | "sterilizer" |
| 3 | "Inphormex" |
| 4 | "sterilizer" |
| 5 | "sterilizer" |
75: modify JSON
select
ident,
name,
json_set(details, '$.sold', json_quote('2024-01-25')) as updated
from machine;
| ident | name | updated |
|-------|----------------|--------------------------------------------------------------|
| 1 | WY401 | {"acquired":"2023-05-01","sold":"2024-01-25"} |
| 2 | Inphormex | {"acquired":"2021-07-15","refurbished":"2023-10-22","sold":" |
| | | 2024-01-25"} |
| 3 | AutoPlate 9000 | {"note":"needs software update","sold":"2024-01-25"} |
- Updates the in-memory copy of the JSON, not the database record
- Please use
json_quoterather than trying to format JSON with string operations
☆ refresh penguins
select species, count(*) as num
from penguins
group by species;
| species | num |
|-----------|-----|
| Adelie | 152 |
| Chinstrap | 68 |
| Gentoo | 124 |
- We will restore full database after each example
76: tombstones
alter table penguins
add active integer not null default 1;
update penguins
set active = iif(species = 'Adelie', 0, 1);
select species, count(*) as num
from penguins
where active
group by species;
| species | num |
|-----------|-----|
| Chinstrap | 68 |
| Gentoo | 124 |
- Use a tombstone to mark (in)active records
- Every query must now include it
77: views
create view if not exists
active_penguins (
species,
island,
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
sex
) as
select
species,
island,
bill_length_mm,
bill_depth_mm,
flipper_length_mm,
body_mass_g,
sex
from penguins
where active;
select species, count(*) as num
from active_penguins
group by species;
| species | num |
|-----------|-----|
| Chinstrap | 68 |
| Gentoo | 124 |
- A view is a saved query that other queries can invoke
- View is re-run each time it’s used
- Like a CTE, but:
- Can be shared between queries
- Views came first
- Some databases offer materialized views
- Update-on-demand temporary tables
☆ hours reminder
create table job (
name text not null,
billable real not null
);
insert into job values
('calibrate', 1.5),
('clean', 0.5);
select * from job;
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
| clean | 0.5 |
78: add check
create table job (
name text not null,
billable real not null,
check (billable > 0.0)
);
insert into job values ('calibrate', 1.5);
insert into job values ('reset', -0.5);
select * from job;
Runtime error near line 9: CHECK constraint failed: billable > 0.0 (19)
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
checkadds constraint to table- Must produce a Boolean result
- Run each time values added or modified
- But changes made before the error have taken effect
☆ ACID
- Atomic: change cannot be broken down into smaller ones (i.e., all or nothing)
- Consistent: database goes from one consistent state to another
- Isolated: looks like changes happened one after another
- Durable: if change takes place, it’s still there after a restart
79: transactions
create table job (
name text not null,
billable real not null,
check (billable > 0.0)
);
insert into job values ('calibrate', 1.5);
begin transaction;
insert into job values ('clean', 0.5);
rollback;
select * from job;
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
- Statements outside transaction execute and are committed immediately
- Statement(s) inside transaction don’t take effect until:
end transaction(success)rollback(undo)
- Can have any number of statements inside a transaction
- But cannot nest transactions in SQLite
- Other databases support this
80: rollback in constraint
create table job (
name text not null,
billable real not null,
check (billable > 0.0) on conflict rollback
);
insert into job values
('calibrate', 1.5);
insert into job values
('clean', 0.5),
('reset', -0.5);
select * from job;
Runtime error near line 11: CHECK constraint failed: billable > 0.0 (19)
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
- All of second
insertrolled back as soon as error occurred - But first
inserttook effect
81: rollback in statement
create table job (
name text not null,
billable real not null,
check (billable > 0.0)
);
insert or rollback into job values
('calibrate', 1.5);
insert or rollback into job values
('clean', 0.5),
('reset', -0.5);
select * from job;
Runtime error near line 11: CHECK constraint failed: billable > 0.0 (19)
| name | billable |
|-----------|----------|
| calibrate | 1.5 |
- Constraint is in table definition
- Action is in statement
82: upsert
create table jobs_done (
person text unique,
num integer default 0
);
insert into jobs_done values("zia", 1);
.print "after first"
select * from jobs_done;
.print
insert into jobs_done values("zia", 1);
.print "after failed"
select * from jobs_done;
insert into jobs_done values("zia", 1)
on conflict(person) do update set num = num + 1;
.print "\nafter upsert"
select * from jobs_done;
after first
| person | num |
|--------|-----|
| zia | 1 |
Runtime error near line 14: UNIQUE constraint failed: jobs_done.person (19)
after failed
| person | num |
|--------|-----|
| zia | 1 |
after upsert
| person | num |
|--------|-----|
| zia | 2 |
- upsert stands for “update or insert”
- Create if record doesn’t exist
- Update if it does
- Not standard SQL but widely implemented
- Example also shows use of SQLite
.printcommand
☆ normalization
-
First normal form (1NF): every field of every record contains one indivisible value.
-
Second normal form (2NF) and third normal form (3NF): every value in a record that isn’t a key depends solely on the key, not on other values.
-
Denormalization: explicitly store values that could be calculated on the fly
- To simplify queries and/or make processing faster
83: create trigger
- A trigger automatically runs before or after a specified operation
- Can have side effects (e.g., update some other table)
- And/or implement checks (e.g., make sure other records exist)
- Add processing overhead…
- …but data is either cheap or correct, never both
- Inside trigger, refer to old and new versions of record
as
old.columnandnew.column
-- Track hours of lab work.
create table job (
person text not null,
reported real not null check (reported >= 0.0)
);
-- Explicitly store per-person total rather than using sum().
create table total (
person text unique not null,
hours real
);
-- Initialize totals.
insert into total values
("gene", 0.0),
("august", 0.0);
-- Define a trigger.
create trigger total_trigger
before insert on job
begin
-- Check that the person exists.
select case
when not exists (select 1 from total where person = new.person)
then raise(rollback, 'Unknown person ')
end;
-- Update their total hours (or fail if non-negative constraint violated).
update total
set hours = hours + new.reported
where total.person = new.person;
end;
insert into job values
('gene', 1.5),
('august', 0.5),
('gene', 1.0)
;
| person | reported |
|--------|----------|
| gene | 1.5 |
| august | 0.5 |
| gene | 1.0 |
| person | hours |
|--------|-------|
| gene | 2.5 |
| august | 0.5 |
081: trigger firing
insert into job values
('gene', 1.0),
('august', -1.0)
;
Runtime error near line 6: CHECK constraint failed: reported >= 0.0 (19)
| person | hours |
|--------|-------|
| gene | 0.0 |
| august | 0.0 |
☆ represent graphs
create table lineage (
parent text not null,
child text not null
);
insert into lineage values
('Arturo', 'Clemente'),
('Darío', 'Clemente'),
('Clemente', 'Homero'),
('Clemente', 'Ivonne'),
('Ivonne', 'Lourdes'),
('Soledad', 'Lourdes'),
('Lourdes', 'Santiago');
select * from lineage;
| parent | child |
|----------|----------|
| Arturo | Clemente |
| Darío | Clemente |
| Clemente | Homero |
| Clemente | Ivonne |
| Ivonne | Lourdes |
| Soledad | Lourdes |
| Lourdes | Santiago |
84: recursive query
with recursive descendent as (
select
'Clemente' as person,
0 as generations
union all
select
lineage.child as person,
descendent.generations + 1 as generations
from descendent join lineage
on descendent.person = lineage.parent
)
select person, generations from descendent;
| person | generations |
|----------|-------------|
| Clemente | 0 |
| Homero | 1 |
| Ivonne | 1 |
| Lourdes | 2 |
| Santiago | 3 |
- Use a recursive CTE to create a temporary table (
descendent) - Base case seeds this table
- Recursive case relies on value(s) already in that table and external table(s)
union allto combine rows- Can use
unionbut that has lower performance (must check uniqueness each time)
- Can use
- Stops when the recursive case yields an empty row set (nothing new to add)
- Then select the desired values from the CTE
☆ contact tracing database
select * from person;
| ident | name |
|-------|-----------------------|
| 1 | Juana Baeza |
| 2 | Agustín Rodríquez |
| 3 | Ariadna Caraballo |
| 4 | Micaela Laboy |
| 5 | Verónica Altamirano |
| 6 | Reina Rivero |
| 7 | Elias Merino |
| 8 | Minerva Guerrero |
| 9 | Mauro Balderas |
| 10 | Pilar Alarcón |
| 11 | Daniela Menéndez |
| 12 | Marco Antonio Barrera |
| 13 | Cristal Soliz |
| 14 | Bernardo Narváez |
| 15 | Óscar Barrios |
select * from contact;
| left | right |
|-------------------|-----------------------|
| Agustín Rodríquez | Ariadna Caraballo |
| Agustín Rodríquez | Verónica Altamirano |
| Juana Baeza | Verónica Altamirano |
| Juana Baeza | Micaela Laboy |
| Pilar Alarcón | Reina Rivero |
| Cristal Soliz | Marco Antonio Barrera |
| Cristal Soliz | Daniela Menéndez |
| Daniela Menéndez | Marco Antonio Barrera |
85: bidirectional contacts
create temporary table bi_contact (
left text,
right text
);
insert into bi_contact
select
left, right from contact
union all
select right, left from contact
;
| original_count |
|----------------|
| 8 |
| num_contact |
|-------------|
| 16 |
- Create a temporary table rather than using a long chain of CTEs
- Only lasts as long as the session (not saved to disk)
- Duplicate information rather than writing more complicated query
86: update group identifiers
select
left.name as left_name,
left.ident as left_ident,
right.name as right_name,
right.ident as right_ident,
min(left.ident, right.ident) as new_ident
from
(person as left join bi_contact on left.name = bi_contact.left)
join person as right on bi_contact.right = right.name;
| left_name | left_ident | right_name | right_ident | new_ident |
|-----------------------|------------|-----------------------|-------------|-----------|
| Juana Baeza | 1 | Micaela Laboy | 4 | 1 |
| Juana Baeza | 1 | Verónica Altamirano | 5 | 1 |
| Agustín Rodríquez | 2 | Ariadna Caraballo | 3 | 2 |
| Agustín Rodríquez | 2 | Verónica Altamirano | 5 | 2 |
| Ariadna Caraballo | 3 | Agustín Rodríquez | 2 | 2 |
| Micaela Laboy | 4 | Juana Baeza | 1 | 1 |
| Verónica Altamirano | 5 | Agustín Rodríquez | 2 | 2 |
| Verónica Altamirano | 5 | Juana Baeza | 1 | 1 |
| Reina Rivero | 6 | Pilar Alarcón | 10 | 6 |
| Pilar Alarcón | 10 | Reina Rivero | 6 | 6 |
| Daniela Menéndez | 11 | Cristal Soliz | 13 | 11 |
| Daniela Menéndez | 11 | Marco Antonio Barrera | 12 | 11 |
| Marco Antonio Barrera | 12 | Cristal Soliz | 13 | 12 |
| Marco Antonio Barrera | 12 | Daniela Menéndez | 11 | 11 |
| Cristal Soliz | 13 | Daniela Menéndez | 11 | 11 |
| Cristal Soliz | 13 | Marco Antonio Barrera | 12 | 12 |
new_identis minimum of own identifier and identifiers one step away- Doesn’t keep people with no contacts
87: recursive labeling
with recursive labeled as (
select
person.name as name,
person.ident as label
from
person
union -- not 'union all'
select
person.name as name,
labeled.label as label
from
(person join bi_contact on person.name = bi_contact.left)
join labeled on bi_contact.right = labeled.name
where labeled.label < person.ident
)
select name, min(label) as group_id
from labeled
group by name
order by label, name;
| name | group_id |
|-----------------------|----------|
| Agustín Rodríquez | 1 |
| Ariadna Caraballo | 1 |
| Juana Baeza | 1 |
| Micaela Laboy | 1 |
| Verónica Altamirano | 1 |
| Pilar Alarcón | 6 |
| Reina Rivero | 6 |
| Elias Merino | 7 |
| Minerva Guerrero | 8 |
| Mauro Balderas | 9 |
| Cristal Soliz | 11 |
| Daniela Menéndez | 11 |
| Marco Antonio Barrera | 11 |
| Bernardo Narváez | 14 |
| Óscar Barrios | 15 |
- Use
unioninstead ofunion allto prevent infinite recursion
88: query from Python
import sqlite3
connection = sqlite3.connect("db/penguins.db")
cursor = connection.execute("select count(*) from penguins;")
rows = cursor.fetchall()
print(rows)
[(344,)]
sqlite3is part of Python’s standard library- Create a connection to a database file
- Get a cursor by executing a query
- More common to create cursor and use that to run queries
- Fetch all rows at once as list of tuples
89: incremental fetch
import sqlite3
connection = sqlite3.connect("db/penguins.db")
cursor = connection.cursor()
cursor = cursor.execute("select species, island from penguins limit 5;")
while row := cursor.fetchone():
print(row)
('Adelie', 'Torgersen')
('Adelie', 'Torgersen')
('Adelie', 'Torgersen')
('Adelie', 'Torgersen')
('Adelie', 'Torgersen')
cursor.fetchonereturnsNonewhen no more data- There is also
fetchmany(N)to fetch (up to) a certain number of rows
90: insert, delete, and all that
import sqlite3
connection = sqlite3.connect(":memory:")
cursor = connection.cursor()
cursor.execute("create table example(num integer);")
cursor.execute("insert into example values (10), (20);")
print("after insertion", cursor.execute("select * from example;").fetchall())
cursor.execute("delete from example where num < 15;")
print("after deletion", cursor.execute("select * from example;").fetchall())
after insertion [(10,), (20,)]
after deletion [(20,)]
- Each
executeis its own transaction
91: interpolate values
import sqlite3
connection = sqlite3.connect(":memory:")
cursor = connection.cursor()
cursor.execute("create table example(num integer);")
cursor.executemany("insert into example values (?);", [(10,), (20,)])
print("after insertion", cursor.execute("select * from example;").fetchall())
after insertion [(10,), (20,)]
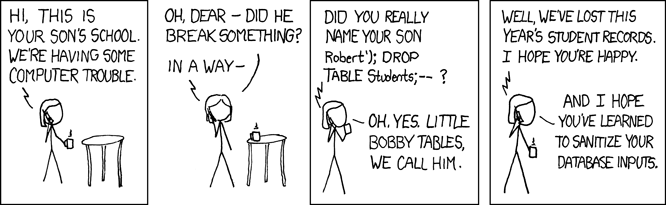
- From XKCD
92: script execution
import sqlite3
SETUP = """\
drop table if exists example;
create table example(num integer);
insert into example values (10), (20);
"""
connection = sqlite3.connect(":memory:")
cursor = connection.cursor()
cursor.executescript(SETUP)
print("after insertion", cursor.execute("select * from example;").fetchall())
after insertion [(10,), (20,)]
- But what if something goes wrong?
93: SQLite exceptions in Python
import sqlite3
SETUP = """\
create table example(num integer check(num > 0));
insert into example values (10);
insert into example values (-1);
insert into example values (20);
"""
connection = sqlite3.connect(":memory:")
cursor = connection.cursor()
try:
cursor.executescript(SETUP)
except sqlite3.Error as exc:
print(f"SQLite exception: {exc}")
print("after execution", cursor.execute("select * from example;").fetchall())
SQLite exception: CHECK constraint failed: num > 0
after execution [(10,)]
94: Python in SQLite
import sqlite3
SETUP = """\
create table example(num integer);
insert into example values (-10), (10), (20), (30);
"""
def clip(value):
if value < 0:
return 0
if value > 20:
return 20
return value
connection = sqlite3.connect(":memory:")
connection.create_function("clip", 1, clip)
cursor = connection.cursor()
cursor.executescript(SETUP)
for row in cursor.execute("select num, clip(num) from example;").fetchall():
print(row)
(-10, 0)
(10, 10)
(20, 20)
(30, 20)
- SQLite calls back into Python to execute the function
- Other databases can run Python (and other languages) in the database server process
- Be careful
95: handle dates and times
from datetime import date
import sqlite3
# Convert date to ISO-formatted string when writing to database
def _adapt_date_iso(val):
return val.isoformat()
sqlite3.register_adapter(date, _adapt_date_iso)
# Convert ISO-formatted string to date when reading from database
def _convert_date(val):
return date.fromisoformat(val.decode())
sqlite3.register_converter("date", _convert_date)
SETUP = """\
create table events(
happened date not null,
description text not null
);
"""
connection = sqlite3.connect(":memory:", detect_types=sqlite3.PARSE_DECLTYPES)
cursor = connection.cursor()
cursor.execute(SETUP)
cursor.executemany(
"insert into events values (?, ?);",
[(date(2024, 1, 10), "started tutorial"), (date(2024, 1, 29), "finished tutorial")],
)
for row in cursor.execute("select * from events;").fetchall():
print(row)
(datetime.date(2024, 1, 10), 'started tutorial')
(datetime.date(2024, 1, 29), 'finished tutorial')
sqlite3.PARSE_DECLTYPEStellssqlite3library to use converts based on declared column types- Adapt on the way in, convert on the way out
96: SQL in Jupyter notebooks
pip install jupysql
- And then inside the notebook:
%load_ext sql
- Loads extension
%sql sqlite:///data/penguins.db
Connecting to 'sqlite:///data/penguins.db'
- Connects to database
sqlite://with two slashes is the protocol/data/penguins.db(one leading slash) is a local path
- Single percent sign
%sqlintroduces one-line command - Use double percent sign
%%sqlto indicate that the rest of the cell is SQL
%%sql
select species, count(*) as num
from penguins
group by species;
Running query in 'sqlite:///data/penguins.db'
| species | num |
|---|---|
| Adelie | 152 |
| Chinstrap | 68 |
| Gentoo | 124 |
97: Pandas and SQL
pip install pandas
import pandas as pd
import sqlite3
connection = sqlite3.connect("db/penguins.db")
query = "select species, count(*) as num from penguins group by species;"
df = pd.read_sql(query, connection)
print(df)
species num
0 Adelie 152
1 Chinstrap 68
2 Gentoo 124
- Be careful about datatype conversion
98: Polars and SQL
pip install polars pyarrow adbc-driver-sqlite
import polars as pl
query = "select species, count(*) as num from penguins group by species;"
uri = "sqlite:///db/penguins.db"
df = pl.read_database_uri(query, uri, engine="adbc")
print(df)
shape: (3, 2)
┌───────────┬─────┐
│ species ┆ num │
│ --- ┆ --- │
│ str ┆ i64 │
╞═══════════╪═════╡
│ Adelie ┆ 152 │
│ Chinstrap ┆ 68 │
│ Gentoo ┆ 124 │
└───────────┴─────┘
- The Uniform Resource Identifier (URI) specifies the database
- The query is the query
- Use the ADBC engine instead of the default ConnectorX
99: object-relational mapper
from sqlmodel import Field, Session, SQLModel, create_engine, select
class Department(SQLModel, table=True):
ident: str = Field(default=None, primary_key=True)
name: str
building: str
engine = create_engine("sqlite:///db/assays.db")
with Session(engine) as session:
statement = select(Department)
for result in session.exec(statement).all():
print(result)
building='Chesson' name='Genetics' ident='gen'
building='Fashet Extension' name='Histology' ident='hist'
building='Chesson' name='Molecular Biology' ident='mb'
building='TGVH' name='Endocrinology' ident='end'
- An object-relational mapper (ORM) translates table columns to object properties and vice versa
- SQLModel relies on Python type hints
100: relations with ORM
class Staff(SQLModel, table=True):
ident: str = Field(default=None, primary_key=True)
personal: str
family: str
dept: Optional[str] = Field(default=None, foreign_key="department.ident")
age: int
engine = create_engine("sqlite:///db/assays.db")
SQLModel.metadata.create_all(engine)
with Session(engine) as session:
statement = select(Department, Staff).where(Staff.dept == Department.ident)
for dept, staff in session.exec(statement):
print(f"{dept.name}: {staff.personal} {staff.family}")
Histology: Divit Dhaliwal
Molecular Biology: Indrans Sridhar
Molecular Biology: Pranay Khanna
Histology: Vedika Rout
Genetics: Abram Chokshi
Histology: Romil Kapoor
Molecular Biology: Ishaan Ramaswamy
Genetics: Nitya Lal
- Make foreign keys explicit in class definitions
- SQLModel automatically does the join
- The two staff with no department aren’t included in the result
☆ Appendices
☆ Terms
- 1-to-1 relation
- A relationship between two tables in which each record from the first table matches exactly one record from the second and vice versa.
- 1-to-many relation
- A relationship between two tables in which each record from the first table matches zero or more records from the second, but each record from the second table matches exactly one record from the first.
- aggregation
- Combining several values to produce one.
- aggregation function
- A function used to produce one value from many, such as maximum or addition.
- alias
- An alternate name used temporarily for a table or column.
- atomic
- An operation that cannot be broken into smaller operations.
- autoincrement
- Automatically add one to a value.
- base case
- A starting point for recursion that does not depend on previous recursive calculations.
- Binary Large Object (blob)
- Bytes that are handled as-is rather than being interpreted as numbers, text, or other data types.
- cross join
- A join that creates the cross-product of rows from two tables.
- common table expression (CTE)
- A temporary table created at the start of a query, usually to simplify writing the query.
- consistent
- A state in which all constraints are satisfied, e.g., all columns contain allowed values and all foreign keys refer to primary keys.
- A subquery that depends on a value or values from the enclosing query, and which must therefore be executed once for each of those values.
- cursor
- A reference to the current location in the results of an ongoing query.
- data migration
- To move data from one form to another, e.g., from one set of tables to a new set or from one DBMS to another.
- database
- A collection of data that can be searched and retrieved.
- database management system (DBMS)
- A program that manages a particular kind of database.
- denormalization
- To deliberately introduce duplication or other violate normal forms, typically to improve query performance.
- durable
- Guaranteed to survive shutdown and restart.
- entity-relationship diagram
- A graphical depiction of the relationships between tables in a database.
- filter
- To select records based on whether they pass some Boolean test.
- foreign key
- A value in one table that identifies a primary key in another table.
- full outer join
- See cross join.
- group
- A set of records that share a common property, such as having the same value in a particular column.
- in-memory database
- A database that is stored in memory rather than on disk.
- index
- An auxiliary data structure that enables faster access to records.
- infinite recursion
- See “infinite recursion”.
- isolated
- The appearance of having executed in an otherwise-idle system.
- join
- To combine records from two tables.
- join condition
- The criteria used to decide which rows from each table in a join are combined.
- join table
- A table that exists solely to enable information from two tables to be connected.
- left outer join
- A join that is guaranteed to keep all rows from the first (left) table. Columns from the right table are filled with actual values if available or with null otherwise.
- many-to-many relation
- A relationship between two tables in which each record from the first table may match zero or more records from the second and vice versa.
- materialized view
- A view that is stored on disk and updated on demand.
- normal form
- One of several (loosely defined) rules for organizing data in tables.
- NoSQL database
- Any database that doesn’t use the relational model.
- null
- A special value representing “not known”.
- object-relational mapper (ORM)
- A library that translates objects in a program into database queries and the results of those queries back into objects.
- path expression
- An expression identifying an element or a set of elements in a JSON structure.
- primary key
- A value or values in a database table that uniquely identifies each record in that table.
- query
- A command to perform some operation in a database (typically data retrieval).
- recursive CTE
- A common table expression that refers to itself. Every recursive CTE must have a base case and a recursive case.
- recursive case
- The second or subsequent step in self-referential accumulation of data.
- relational database management system (RDBMS)
- A database management system that stores data in tables with columns and rows.
- subquery
- A query used within another query.
- table-valued function
- A function that returns multiple values rather than a single value.
- temporary table
- A table that is explicitly constructed in memory outside any particular query.
- ternary logic
- A logic based on three values: true, false, and “don’t know” (represented as null).
- tombstone
- A marker value added to a record to show that it is no longer active. Tombstones are used as an alternative to deleting data.
- trigger
- An action that runs automatically when something happens in a database, typically insertion or deletion.
- upsert
- To update a record if it exists or insert (create) a new record if it doesn’t.
- Uniform Resource Identifier (URI)
- A string that identifies a resource (such as a web page or database) and the protocol used to access it.
- view
- A rearrangement of data in a database that is regenerated on demand.
- window function
- A function that combines data from adjacent rows in a database query’s result.
☆ Acknowledgments
This tutorial would not have been possible without:
- Andi Albrecht’s
sqlparsemodule - Dimitri Fontaine’s The Art of PostgreSQL
- David Rozenshtein’s The Essence of SQL (now sadly out of print)
I would also like to thank the following for spotting issues, making suggestions, or submitting changes:
- Sam Hames
- Robert Kern
- Roy Pardee
- Manos Pitsidianakis
- Daniel Possenriede
- Adam Rosien
- Thomas Sandmann
- Simon Willison









